Assembling a text-mining corpus for the Ras pathway
Posted on May 19, 2016 in text mining
Over the last year and a half or so I've been involved in the Big Mechanism program sponsored by DARPA. The practical goal of this program is to develop software systems to extract facts from the scientific literature by text mining and, from these facts, assemble causal, mechanistic models that can be used to explain and predict phenomena. The bigger picture goal is to explore an approach to science in which machines assume a greater share of the burden in aggregating and integrating research. Though DARPA envisions applications of this type of technology in multiple domains, the initial focus of the Big Mechanism program is in cancer biology, specifically Ras-driven cancer.
Most of my work on the Big Mechanism program up to this point has been to develop tools that assemble mechanisms into models, deconflicting, cleaning, and assembling findings into different formats. Having had some success in automated assembly of signaling models from databases (such as Pathway Commons) we are now looking to see how much more we can enrich these models using large-scale machine reading.
I started looking into this in the context of a specific use case: assembling a large-scale, high-quality model of the Ras signaling pathway, which I've been developing along with Ben Gyori, Kartik Subramanian and other collaborators here at HMS. As a starting point, we've defined the Ras signaling pathway according to Frank McCormick's RAS Pathway v2.0 diagram and accompanying table, which includes 227 genes organized into 65 groups.
The first question that arises is what is the best way to find papers relevant to a set of genes? A requirement is that the process of querying for publications should be automated, with minimal human intervention or curation. I tried two (very simple) approaches:
- Query Pubmed using the canonical (HGNC) gene name
- Get the set of references associated with the gene from the Entrez Gene database.
The first approach produces a ton of results, but has some issues: for one, it picks up false positives due to gene names that may be matches to other things: for example, the gene name JUN seems to pick any paper published in the month of June. On the other hand, this approach also seems to miss relevant papers due to the fact that most genes have several synonyms and many papers may refer to the gene using non-standard names.
The second approach pulls the curated PMID references associated with the gene from the Entrez Gene database. The set of PMIDs obtained by pulling all PMIDs out of the XML result for the gene corresponds closely to the "Bibliography" section of the Entrez Gene information page (e.g., see the Bibliography section for BRCA1).
As expected, searching by gene name returns a much larger set of PMIDs (more than 6 times larger) than obtaining the references from Entrez Gene. In both cases there was a substantial fraction of papers that were returned by searches for multiple genes, as might be expected for genes identified a priori as being involved in a common biological process. In both cases roughly 75% of the assembled list of PMIDs were unique.
| By gene name | By gene ID | |
|---|---|---|
| Total refs | 464,917 | 74,529 |
| Unique refs | 355,781 | 54,308 |
How many citations do we tend to get by gene? The figure below shows the distribution of the number of PMIDs returned for each gene, sorted by the number of PMIDs returned by gene name search, and plotted on a log scale. The distribution roughly follows a power law, with deviations for the most-cited and least-cited genes.
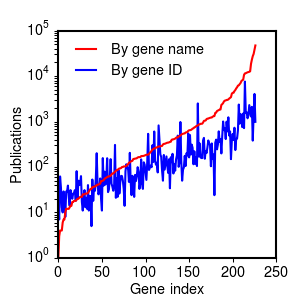
Citation distribution for 227 Ras genes, sorted by citation count for name-based search (PDF).
Reassuringly, the number of references returned by the gene ID search roughly follows the number of references returned by the name search, but with substantially fewer references overall. The least-cited genes appear to be an exception to this pattern: for these the gene ID search appears to return a larger number of references than the name search. This appears to be due to the fact that the least-cited genes often appear in the literature under different names, and Entrez Gene collates citations across multiple names.
The list of the top 10 genes (by citations) returns reassuringly familiar names. If anything, the gene ID search returns a list closer to what one might expect from how "famous" the genes tend to be, suggesting that it's less susceptible to variability due to the use of the particular name in the literature. For example, it's surprising that TP53 doesn't make the top 10 in the gene name search, probably because it's more frequently referred to by its protein name, p53, than its official gene name, TP53. Similarly, FOS is number 4 on the gene name list, but it's certainly not as well known as NFKB1 or KRAS, both of which make the top 10 by gene ID but not by gene name. A quick scan of the search results for "FOS" revealed hits not only for the gene FOS, but also false positives like "fructooligosaccharide" (FOS), "Framingham Offspring Study" (FOS), and "foot orthoses" (FOs).
| Rank | By gene name (refs) | By gene ID (refs) |
|---|---|---|
| 1 | CASP3 (47320) | TP53 (7598) |
| 2 | EGFR (38072) | EGFR (4056) |
| 3 | MYC (30819) | NFKB1 (2508) |
| 4 | FOS (27521) | AKT1 (2370) |
| 5 | ERBB2 (23076) | BRCA1 (2304) |
| 6 | MTOR (18677) | ERBB2 (2107) |
| 7 | MAPK1 (12766) | MAPK1 (1719) |
| 8 | BRCA1 (12458) | KRAS (1609) |
| 9 | CDKN1A (12266) | PTEN (1571) |
| 10 | MAPK3 (12144) | BRAF (1503) |
The genes with the fewest citations have a surprisingly small number of references given that they were explicitly included in a curated set of key Ras pathway genes. Many of them are lesser-known isoforms of widely-studied gene families (e.g., SPRED3, RASA2, PIK3R5/6):
| Rank | By gene name (refs) | By gene ID (refs) |
|---|---|---|
| 218 | PIK3R5 (12) | RALGAPA2 (13) |
| 219 | SPRED3 (10) | RASGRP4 (13) |
| 220 | EXOC1 (7) | SPRY3 (12) |
| 221 | RALGAPA1 (7) | RASA2 (11) |
| 222 | RASSF9 (6) | RASSF10 (11) |
| 223 | CYTH2 (4) | RGL1 (11) |
| 224 | EXOC6 (4) | RASSF9 (10) |
| 225 | RALGAPA2 (4) | SPRED3 (8) |
| 226 | RASAL3 (3) | RASAL3 (7) |
| 227 | PIK3R6 (1) | RGL3 (5) |
There are of course many other ways to assemple corpora, including systematic use of gene synonyms, exploiting MeSH terms and other metadata, as well as using other search tools (e.g., CrossRef). These were two very simple ways to get a sense of the scale of the relevant literature, with an expansive and a restricted search giving rough upper and lower bounds. My conclusion is that the curated references in Entrez Gene are less likely to contain false positives, with the downside of missing many potentially relevant articles. Given that the size of the corpus returned by Entrez Gene search is smaller, I'll use this set of roughly ~54k papers for an initial pilot study in machine reading for mechanisms.
In a subsequent post, I'll look at what fraction of the articles in these two corpora are available for text mining from Pubmed Central.