We need to talk about how we talk about protein families and complexes
Posted on July 08, 2018 in Big Mechanism, INDRA
(Our paper on FamPlex, a semantic resource for improving text mining and biocuration for protein families and complexes, is available at BMC Bioinformatics here. Synopsis follows below.)
When Ben Gyori and I started working with natural language processing (NLP) systems as part of the DARPA Big Mechanism program, we found that all the systems we worked with had a common problem: they were frequently unable to correctly and uniformly identify common protein families and complexes.
When I say "identify," I mean assign a database identifier (e.g., Uniprot ID, HGNC ID, Gene Ontology ID, etc.) to a text string denoting a protein family or complex (e.g., NF-kappaB, a complex, or Ras, a gene family). In the NLP world, this process is called "named entity linking", "named entity normalization", or simply "grounding."
Why is this important? Like many others in the systems/computational biology community, we are interested in mining the scientific literature for mechanistic information that can we can use to analyze data and build predictive, explanatory models. The problem in a nutshell is that scientists often write and talk about biological mechanisms in terms of protein families and functional complexes, whereas biological datasets are invariably expressed in terms of the abundances or activities of specific genes or proteins. So if we are going to make use of the (large!) amount of information expressed in terms of families and complexes we have to
- Recognize and ground these terms to standard identifiers, and
- Link these family/complex identifiers to their gene/protein-level constituents.
So why is this difficult for NLP algorithms in practice? Having looked at the types of errors made by two different reading systems, REACH (developed by Mihai Surdeanu's group at the University of Arizona) and TRIPS (developed by the Institute of Human and Machine Cognition), we concluded that the problem was not primarily with the NLP systems and their grounding algorithms, but rather in the lack of uniform resources for grounding and linking families and complexes.
For example, sometimes family/complex entities in text came back from the machine readers with no associated identifiers at all. The reason was usually straightforward: the databases indexed by the reading systems for grounding either didn't have much coverage of families and complexes, or those databases lacked the lexical synonyms necessary for accurate matching. For example, REACH, which indexed Uniprot, InterPro, Pfam, HMDB, ChEBI, Gene Ontology, MeSH, and other ontologies, found no grounding for the string NFkappaB, one of the most frequently occurring in our corpus. In fact, in a corpus of ~215,000 articles (a mix of full texts and abstracts), we found that the seven of ten of the most frequently occurring ungrounded entities were families and complexes!
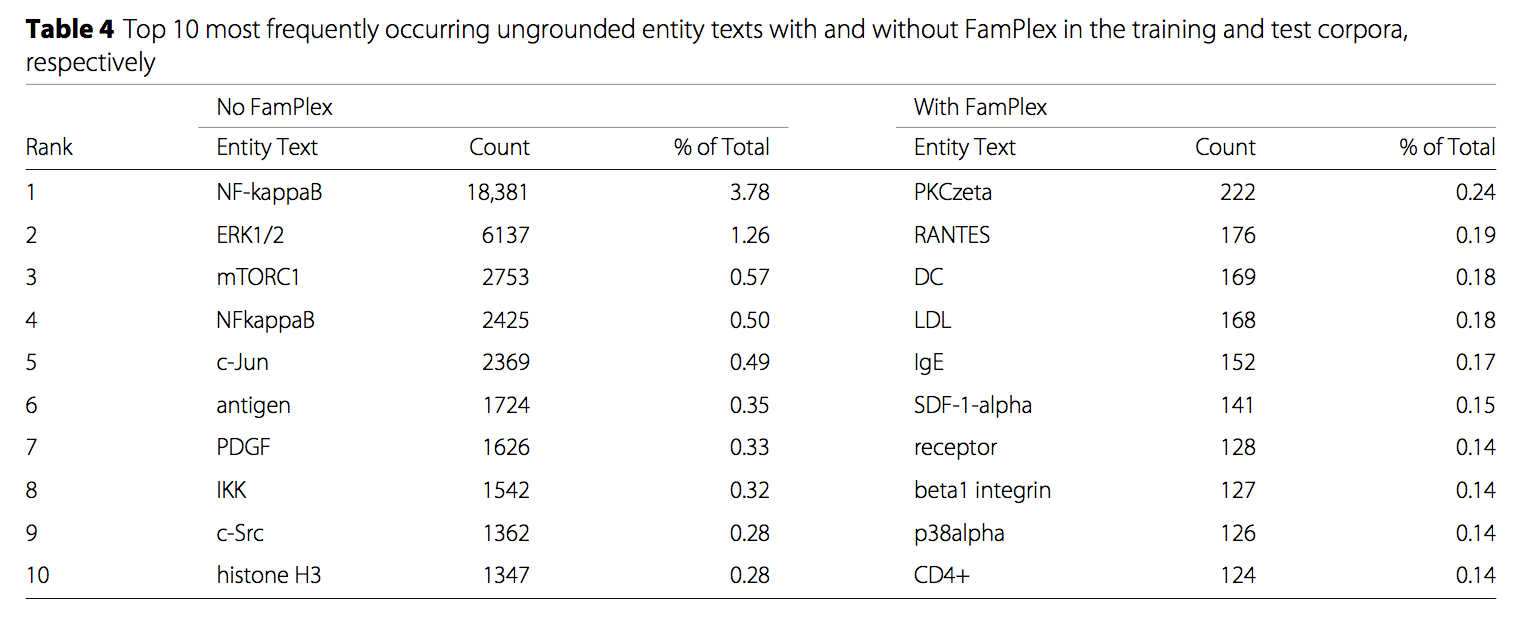
Most frequently occurring ungrounded entity texts, with and without FamPlex. Families and complexes ungrounded without FamPlex are NF-kappaB, ERK1/2, mTORC1, NFkappaB, PDGF, IKK, and histone H3).
In other cases, family/complex names were incorrectly grounded to specific genes due to spurious exact matches in unexpected places. For example, ERK, the common name for the MAPK1/MAPK3 gene family, was incorrectly grounded to EPHB2 due to ERK being listed in Uniprot as a synonym for that gene. Similarly, human gene families were sometimes grounded to the single ortholog of the family in a different organism. My personal favorite was the grounding of AKT to the Dictyostelium (slime mold) gene pkbA instead of the human gene family consisting of the human genes AKT1/AKT2/AKT3.

Uniprot entry for the Dictyostelium gene pkbA has "akt" as a synonym, causing a spurious match.
In our evaluations, we found that the TRIPS system did a bit better, finding a higher percentage of matches using a different matching algorithm and a different set of databases, in particular the NCI Thesaurus (NCIT) and NextProt. Here, though, we found problems with resolving relationships to specific genes: 41% of the entities grounded to NCIT did not have any gene members defined, making it difficult to use this information in downstream data analysis.
More generally, among NLP event extraction systems and biocuration projects there seemed to be a complete lack of consistency in the resources used to identify families and complexes. Among the sources we encountered, there was literally no overlap!
| Source | Type | Family/Complex Databases |
|---|---|---|
| REACH | NLP | InterPro, Pfam, GO |
| TRIPS | NLP | NextProt, NCIT |
| MedScan | NLP | Medscan IDs, Enzyme codes |
| TEES | NLP | Homologene |
| BEL | Curation | Selventa families and complexes |
| Pathway Commons | Curation | (genes enumerated) |
| SIGNOR | Curation | SIGNOR families and complexes |
| Reactome | Curation | Reactome families and complexes |
| EMBO Sourcedata | Curation | Ungrounded or genes enumerated |
This was a problem for us, because we were interested primarily in aggregating and assembling information from both text mining and curated databases.
So...we started curating identifiers ourselves, defining IDs for protein families and complexes, linking in the synonyms that we were seeing in the literature, mapping them to the identifiers in both the protein databases and pathway resources, and defining the hierarchical relationships between complexes, families and their members. Mindful of how standards proliferate, our goal was not to supplant the existing resources but instead provide an extensible "bridging resource" for NLP developers and biocurators to ground and link the most commonly occurring entities and thereby combine information from multiple sources.
The project grew from a handful of CSV files intended for internal use to a fairly robust resource that improved grounding significantly enough that several of the NLP teams in the Big Mechanism program started to use it. We named it "FamPlex", for "Families, Complexes and their Lexicalizations". It consists of a set of files specifying identifiers for 441 human protein families and complexes, their synonyms, gene-level constituents, and equivalent identifiers in other resources.
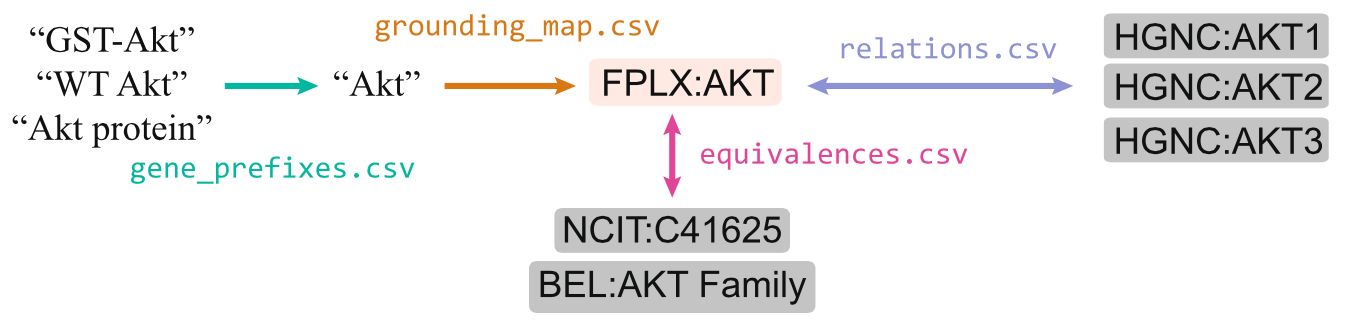
Structure of the FamPlex resource.
We also included a curated list of gene/protein affixes annotated with their semantic meaning, useful for normalizing entity names and correctly interpreting extracted events.
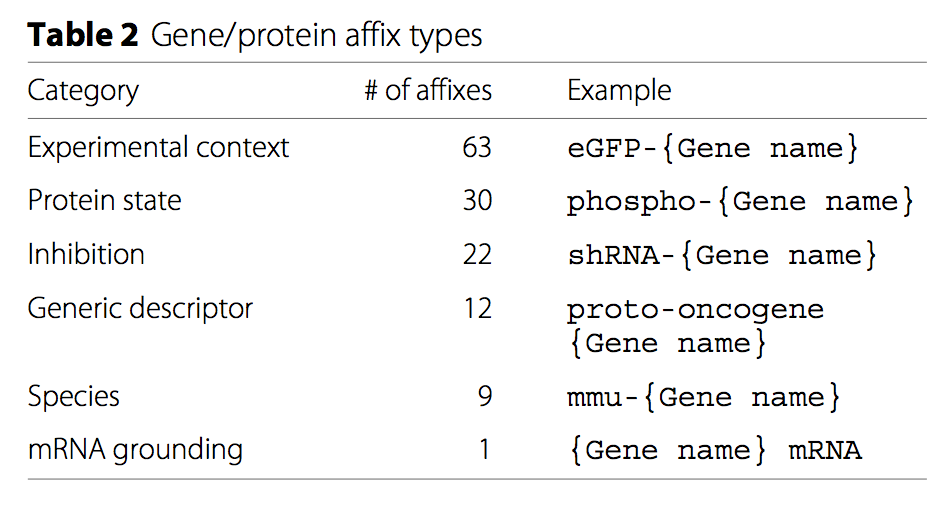
Gene/protein prefixes in FamPlex.
We realized fairly early on that a simple two-layer mapping between families/complexes and their members would not be sufficient to capture the range of entities described in the literature. Articles frequently referred not just to families/complexes and their gene-level members, but often to intermediate groupings of entities, such as a class of subunits forming a part of a functional complex. To handle this, we defined two relationships, isa and partof, that could be nested to define the relationships within a family/complex, as for example with AMPK (a heterotrimer consisting of different combinations of genes drawn from three subunit families, alpha, beta, and gamma). We further defined synonyms for each element in the hierarchy to help NLP systems extract information about mechanisms at any level.
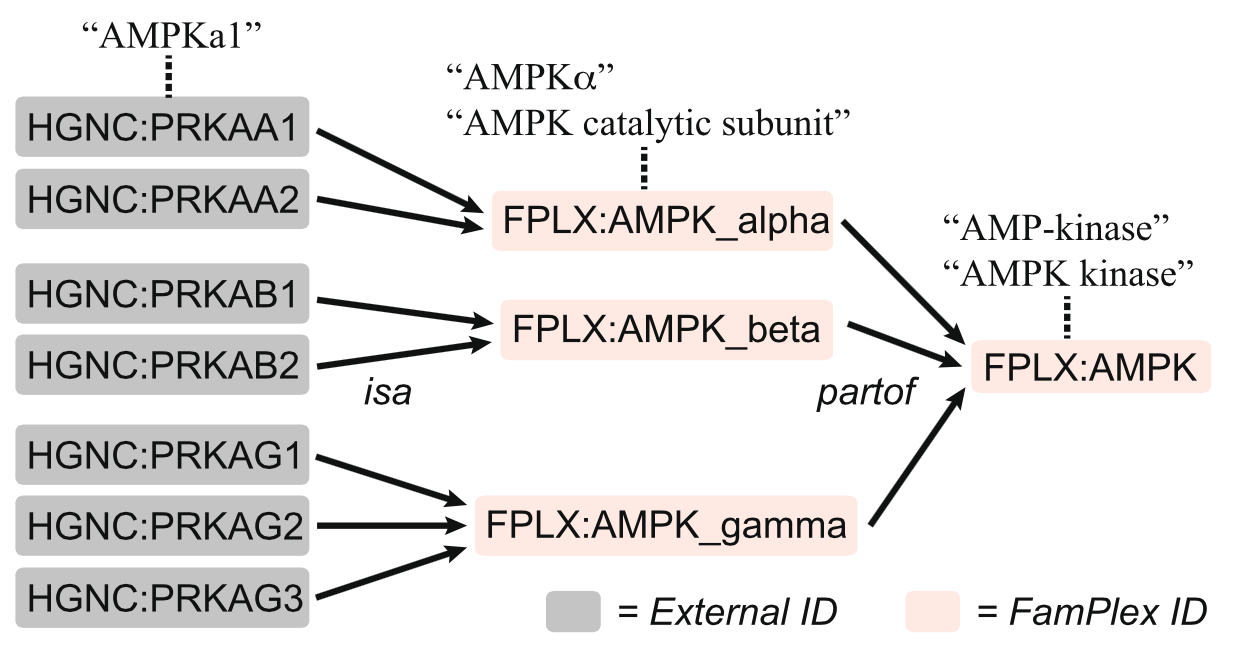
Hierarchical structure of FamPlex relationships, shown here for AMPK.
In sharing and publishing this resource with the community we sought to quantify the improvements attributable to the incorporation of FamPlex in an NLP or biocuration setting. We compared grounding accuracy for two readers (TRIPS and REACH) compiled with and without FamPlex. For REACH, the performance improvement was especially significant, with accuracy for protein families and complex rising from 15% to 71%.
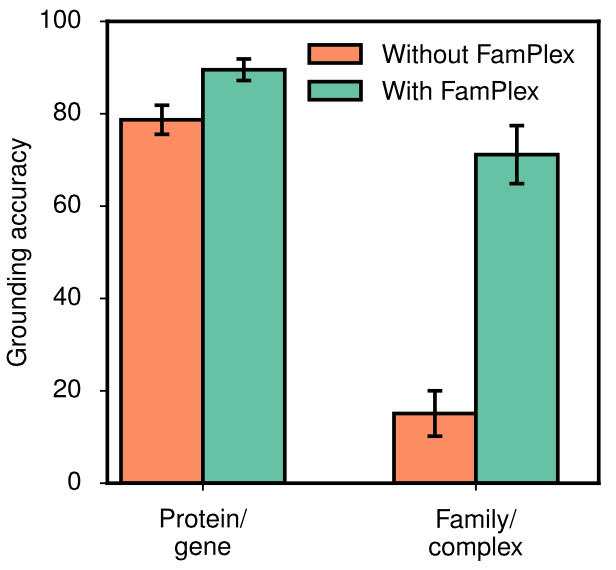
Improvements in grounding accuracy for proteins/genes and families/complexes, with and without the use of FamPlex.
To get an estimate of the coverage of the resource relevant to a real-world biocuration project, we also quantified the proportion of family/complex-level annotations curated in the EMBO Sourcedata dataset that had relevant identifiers in FamPlex and obtained figures of 80-90%.
We also found empirically that the hierarchical structure of FamPlex allowed us to identify relationships between events extracted from families/complexes, genes, and intermediate levels. For some entities, for example Activin (a class of protein complexes belonging to the TGF-beta superfamily), the intermediate levels of representation (e.g., Activin A, Activin B, and Activin AB) were more frequently mentioned in extracted events than either genes or the top-level category!
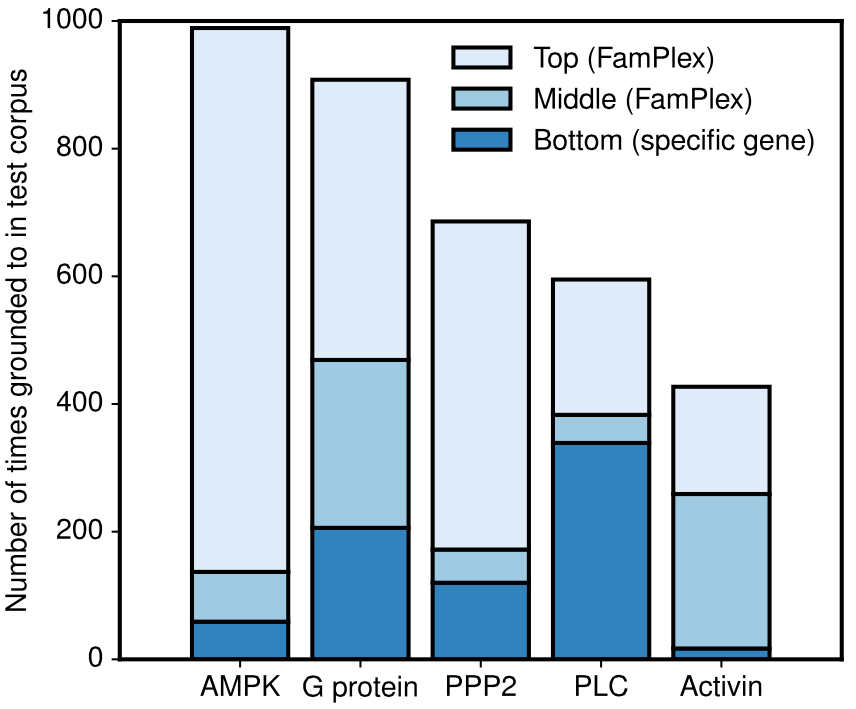
The proportion of groundings at gene-level, intermediate-level, or top-level entities for five-multi-level families/complexes in FamPlex.
Amid a push to facilitate data integration and standardize nomenclature for human genes in the published literature, we speculate that FamPlex could be used to annotate text and data dealing with functional complexes and gene families (e.g., the results of antibody-based experiments involving multiple family members).
For more on the details of the construction and evaluation of FamPlex, see the paper! We've made FamPlex available under a CC0 license so that people can extend, remix, and combine FamPlex with other resources as necessary. We hope that this will be a useful tool for the computational biology and biocuration community. Feel free to ask questions or suggest additions via the Issues page on Github.
Links:
- Paper: https://bmcbioinformatics.biomedcentral.com/articles/10.1186/s12859-018-2211-5
- Code (GitHub): https://github.com/sorgerlab/famplex
- Code for paper: https://github.com/sorgerlab/famplex_paper
- Browse FamPlex at NCBO BioPortal: http://purl.bioontology.org/ontology/FPLX
- identifiers.org prefix: fplx