What fraction of articles can you expect to be available for text mining from Pubmed Central?
Posted on May 20, 2016 in text mining
In a previous post, I described assembling sets of Pubmed references relevant to 227 genes in the Ras pathway. The next problem was getting access to mineable content.
The most readily available source of content for text mining by researchers is the Pubmed Central Open Access article subset, and most demonstrations of text mining tools I have seen within the Big Mechanism program have used this set of articles almost exclusively. These articles have licenses allowing access and reuse suitable for our own (non-commercial research) purposes, though beyond those uses what is permitted depends on the specific, article-by-article license.
For those new to text-mining, as I am, it's worth noting that just because you can read all 4 million full-text articles on the Pubmed Central website doesn't mean you can mine them. In fact, only about ~1.2 million of the articles in Pubmed Central are in the Open Access subset--the majority are off-limits to bulk downloading and mining due to copyright restrictions. And that's out of a total of more than 25 million articles in Pubmed. So depending on what's in your denominator, the fraction of open access articles relevant to your topic could be pretty small.
Another source of articles for text mining is Pubmed Central's Author's Manuscript Collection. These consist of accepted manuscripts uploaded by authors in compliance with the NIH Public Access Policy, and they are also eligible for text mining. This dataset currently consists of ~386,000 articles. This brings the total number of mineable articles from Pubmed central to ~1.6 million.
Some publishers are beginning to make the full texts of non-open-access articles available to subscribers via the CrossRef text and data mining API, which is a subject for another day. Nevertheless, by far the most straightforward way to gain access to the full text of an article for mining is from Pubmed Central--if it's in the Pubmed Central Open Access or Author's Manuscript subsets.
I downloaded the Open Access and Author's Manuscript subsets from PMC and cross-referenced the available content to the full list of PMC articles. Before getting into the fraction of articles in these subsets, it's worth noting that many articles in PMC do not have PMIDs or DOIs associated with them, as shown in the following Venn diagram:
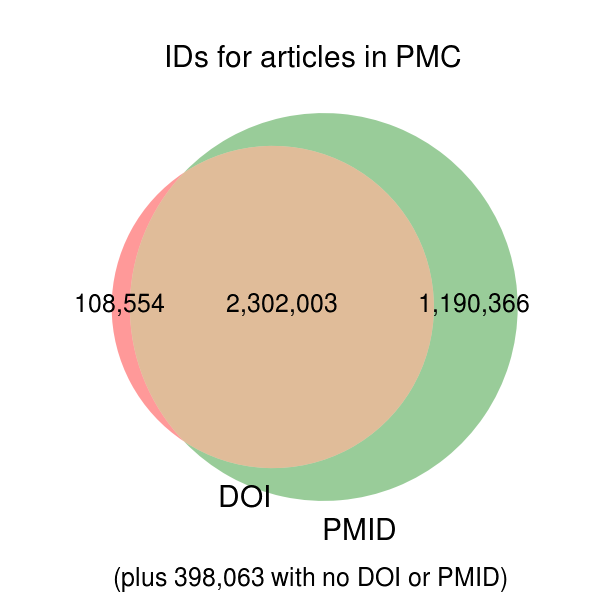
IDs (in addition to PMC ID) associated with the 3,998,986 articles in Pubmed Central. (PDF0)
This has two important consequences: first, since our current approach to building a corpus of papers involves searching Pubmed, we will always be starting with PMIDs, so the ~500,000 articles in Pubmed Central with no PMID are not relevant. Second, for papers that don't turn out to be available in mineable form from PMC, we'll have to look elsewhere (i.e., CrossRef) for full text content, for which we'll need DOIs. Since PMC doesn't have DOIs on file for nearly a third of its articles, we have to obtain them from somewhere else.
Starting with the subset of ~3.5 million articles in PMC that have PMIDs, I looked at how many of these are in the Open Access subset, the Author's Manuscript collection, or both. As the Venn diagram shows, these two collections of mineable articles are mostly complementary, with a relatively small number of articles appearing in both sets.
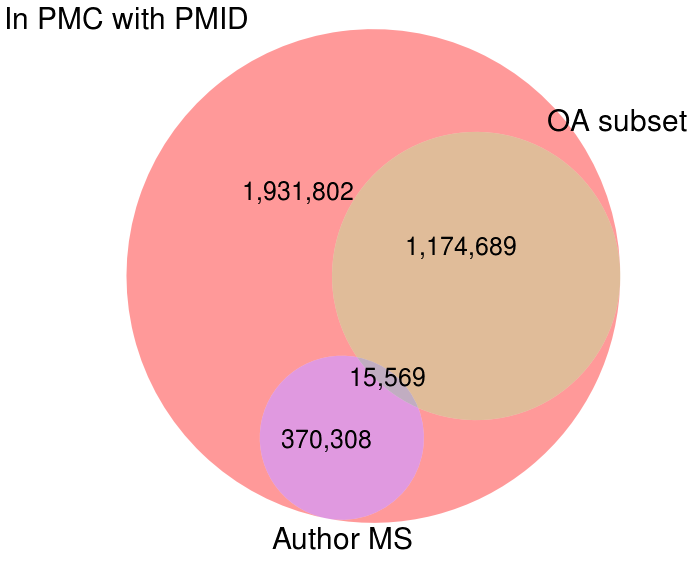
Articles in Pubmed Central that have PMIDs and are available for mining in either the Open Access subset (OA subset) or the Author's Manuscript collection (Author MS). (PDFz)
Next, I looked at the fraction of mineable articles in PMC (OA subset or Author's MS) that one tends to get from Pubmed search results, using the two sets of references for the 227 Ras genes I described in my previous post as examples (a set of ~356,000 papers obtained by gene name searches, and smaller set of ~54,000 papers obtained from the Entrez Gene database).
The results for the larger corpus of 356k papers show a roughly consistent fraction of mineable papers in PMC that appears to be independent of the total number of citations for the gene:
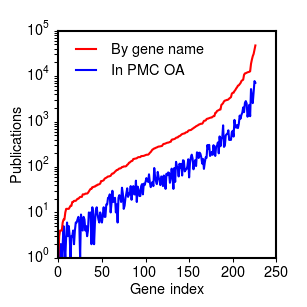
Percentage of references for each gene search with full text in Pubmed Central, sorted by number of total references (PDF1).
The results for the smaller corpus resulting from searching Entrez Gene by gene ID were similar:
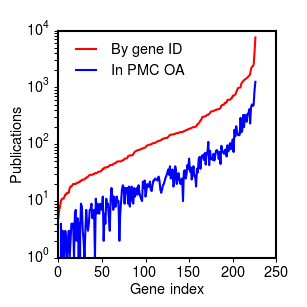
Percentage of references for each gene search with full text in Pubmed Central, sorted by number of total references (PDF2).
For unique references combined across all 227 gene searches, the fraction of mineable articles in Pubmed Central made up roughly 16-18%:
| By gene name | By gene ID | |
|---|---|---|
| Unique refs | 355,781 | 54,308 |
| Mineable In PMC | 58,719 | 9,885 |
| Percentage | 16.5% | 18.2% |
There is another way to look at the data, which is not across the set of references for all genes, but on a gene-by-gene basis. In other words, if you are working on a particular gene, what fraction of the articles on your gene can you expect to find on PMC in a mineable form?
Here is the distribution for the 227 Ras gene searches by gene name:
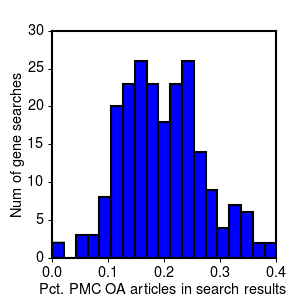
Distribution of full text ratios for different gene name searches (PDF3).
And for the searches in Entrez Gene by gene ID:
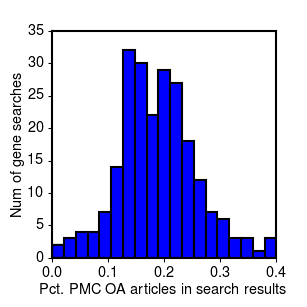
Distribution of full text ratios for references in Entrez gene (PDF4).
The mean and standard deviation for full text percentages across the full set of genes:
| By gene name | By gene ID | |
|---|---|---|
| Mean % in PMC | 20.8% | 18.7% |
| Std Deviation | 9.4% | 6.9% |
As the histograms and the summary statistics show, while on average you might expect to find 1 out of every 5 articles on your gene available for mining in PMC, there is a lot of variability. If you're unlucky, you could easily end up with less than 1 in 10. Moreover, these results are specifically for a set of gene-based searches related to cancer biology. Our experience in obtaining references for two other less molecularly-focused projects (in diabetes and drug-induced cardiotoxicity), suggests that in other domains of biology, the fraction of open access articles may be substantially less, possibly due to different journal or publication practices.